- Home >
- Institut Curie News >
- UNDERSTAND THE ARCHITECTURE AND ORGANIZATION OF CENTROMERES
The work described in this publication, which appeared March 22, 2022 in Molecular Cell, provides implications on the importance of DNA topology for centromeric regulation and stability, resembling those of other repetitive regions of the human genome, such as telomeres and transposons.
The organization of human chromosomes is widely studied and particular attention is paid to the role of repetitive DNA. Much research focuses on centromeres, the well-known chromosomal structures that guide the correct segregation of the genome during mitosis and meiosis. Centromeres are epigenetically regulated structures with DNA repeats in many species. These unique sequences raise questions about the architecture and functionality of centromeric repeats.
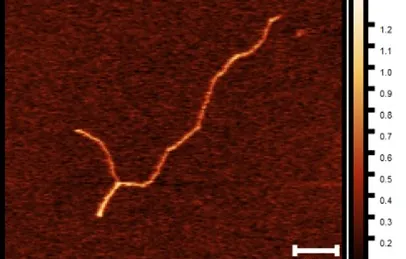
Our paper provides the first report on the complex structures of human centromeric DNA, which regulate centromere positioning and integrity. This work is a team effort where we used interdisciplinary approaches, such as biochemistry, single-molecule techniques (AFM, nanopore, and optical tweezers), and cell imaging, in collaboration with world leaders in single-molecule methods from the University of Amsterdam (Wuite’s team) and Delft University of Technology (Dekker’s team) in the Netherlands. The results of this study are very promising,
explains Daniele Fachinetti, CNRS research director and head of the Molecular Mechanisms of Chromosome Dynamics team in the Cell Biology and Cancer unit (CNRS UMR 144 / Sorbonne University / PSL) at the Institut Curie Research Center.
The team provides direct evidence that human centromeric DNA self-organizes into secondary DNA structures, mainly hairpins of nanometric size. Their work reveals that binding of centromeric protein B (CENP-B) to unique sequences in human centromeric repeats remodels centromeres. This binding forms submicron-sized DNA loops between DNA repeats. These compact the centromeric chromatin and promote inter-chromosome interactions by forming clusters between chromosomes. This is an important feature for preserving centromere positioning in interphase and for maintaining DNA integrity during microtubule pulling during mitosis.
Our results have important implications not only for our understanding of DNA topology and centromeric regulation, but also on genome stability as centromeres or their surrounding regions are hotspots for DNA breakage, but their causes are unknown,
enthuses the molecular biologist.
The surprising discovery of such structures opens the way to future work on the integrity of these regions, for example during replication. Their fragility may indeed be at the root of chromosomal rearrangements observed in several types of cancer.
The formation of DNA loops also reveals a unique and novel high-level organization, which could play a role in maintaining the identity and position of centromeres.
Finally, we highlight a new factor responsible for eukaryotic genome organization: the ability of CENP-B to bring together multiple centromeres. This could have a functional relevance to separate certain centromeres and control DNA metabolism,
concludes the researcher.
► Publication : https://doi.org/10.1016/j.molcel.2022.02.032
CENP-B-mediated DNA loops regulate activity and stability of human
centromeres
Florian Chardon1,#, Aleksandre Japaridze2,#, Hannes Witt3, Leonid Velikovsky1, Camellia
Chakraborty1, Therese Wilhelm1, Marie Dumont1, Wayne Yang2, Carlos Kikuti1, Stephane
Gangnard1, Anne-Sophie Mace1, Gijs Wuite3, Cees Dekker2 and Daniele Fachinetti1,*
1Institut Curie, PSL Research University, CNRS, UMR 144, 26 rue d’Ulm, F-75005, Paris, France
2Department of Bionanoscience, Kavli Institute of Nanoscience Delft, Delft University of
Technology, Van der Maasweg 9, 2629 HZ, Delft, The Netherlands
3Department of Physics and Astronomy, and LaserLaB Amsterdam, Vrije Universiteit
Amsterdam, De Boelelaan 1081, 1081 HV, Amsterdam
#These authors contributed equally to this work
*Corresponding and lead author: daniele.fachinetti@curie.fr