- Home >
- Institut Curie News >
- Spatial transcriptomics: a new guide to better study the spatiality of cells in tissues
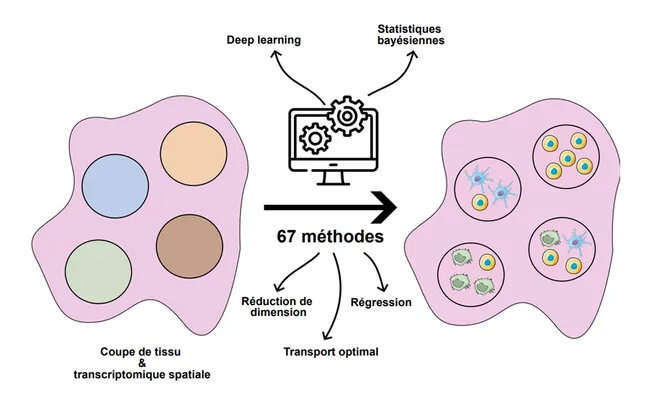
There are many methods that serve to retrieve the spatial distribution of tissue cells from spatial transcriptomic data that do not have single-cell resolution. A team from Institut Curie, Computational Biology and Integrative Genomics of Cancer (Inserm U1331), led by Dr. Florence Cavalli, has recently reviewed all of these analysis methods, known as deconvolution, thus guiding users towards the most appropriate one. This work has been published in Nature Reviews Genetics on May 14, 2025.
Multicellular tissues are made up of different cell types, ensuring their proper functioning within the body. An essential parameter to take into account to understand the functioning of tissues is their spatial organization, i.e. the location of the different cell types within the tissue. Indeed, these are not necessarily distributed homogeneously and do not interact with all other cell types. In organs such as the brain, cell types such as neurons are precisely located in different layers, to ensure the functions of perception, and even memory.
There are several methods for characterizing cells, including the study of their genetic expression via the capture of RNAs (also called transcripts), called transcriptomics. New technologies now make it possible to characterize these cells in their spatial context, and this is called spatial transcriptomics. However, the low resolution of these technologies makes it difficult to accurately assign transcripts to their cell of origin.
" Today there are many ways to analyze the results of spatial transcriptomics that make it possible for us to associate the transcripts with their location in the tissue studied, i.e. to determine from which cells the transcripts come, for example. "explains Florence Cavalli.
In fact, there are 67 so-called deconvolution analysis methods that make it possible to describe where the different cell types are located in a tissue based on spatial transcriptomic data. These methods make it possible to describe the spatial organization of healthy tissues, as well as of tumors. They are described and classified in a new publication of Nature Reviews Genetics, led by the team of Florence Cavalli.
If someone is looking to start this type of analysis today, they will find 67 different methods. Our job here has been to identify all the methods and summarize their common points and their differences in order to guide the user towards the right choice, depending on the biological question, or the potential presence of other data, such as imaging data, which may influence the necessary analysis, "continues to explain Florence Cavalli.
By gathering and comparing all existing analytical methods, this work constitutes a valuable guide to direct users in choosing the most appropriate approach to their research question. Widely used in the fields of oncology, some of these spatial transcriptomic data analysis approaches are used, in particular, in the laboratory of Dr. Florence Cavalli to study the role of cell niches in adult and child brain tumors.
References: Lucie C. Gaspard-Boulinc, Luca Gortana, Thomas Walter, Emmanuel Barillot & Florence M. G. Cavalli. Cell-type deconvolution methods for spatial transcriptomics. Nature Reviews Genetics. May 14, 2025025