- Home >
- Institut Curie News >
- Protein, diversity and evolution
The team of Dr. Sebastian Amigorena, Immune Responses to Cancer (Inserm U932), has for the first time combined protein and evolutionary studies in order to characterize the diversity of more than a thousand proteins integrating transposable element domains, not listed. These results, published in Cell on December 11, 2024, open the way to a better understanding of the functions of proteins in cells, in physiological and pathological conditions.
A well-established dogma in biology states that DNA is transcribed into RNA, which in its turn is translated into proteins. Alternative splicing is a mechanism for bringing diversity to proteins. Simply put, it is the excision and ligation of RNA segments. Thus, the expression of the same gene can, thanks to alternative splicing, produce different protein sequences. The various forms of the same protein are called isoforms.
Unknown isoforms
"Our team, led by Dr. Sebastian Amigorena (Inserm U932), has described a population of alternative splices involving transposable elements, genetic sequences capable of moving to different places in the genome. The isoforms containing these transposable elements are very poorly expressed, and for a long time, the community thought that these were errors, or background noise. We have already described that they are capable of inducing immune responses, but we have now observed that they can also have functions in the body," explains Dr. Yago Arribas De Sandoval, post-doctoral researcher in Dr. Sebastian Amigorena's team and first author of the study.
In addition to being involved in immune processes, the researchers observe that these sequences are present in many individuals and thus hypothesize that these isoforms are not simple errors in protein synthesis, but that they rather serve a role in the cell. By combining transcriptomic study (messenger RNA study) and proteomic study (protein study), they demonstrate that these isoforms are different from the majority isoforms that are called canonical, from which they derive.
New isoforms, new functions?
In order to study the functions of these isoforms in the cell, the researchers selected some of them to reexpress them and study them in detail.
"We have found that they are as stable as canonical proteins and that they localize in specific subcellular compartments, sometimes different from those of canonical isoforms. For example, a stable isoform of a histone (H2AFY) binds to chromatin, even during cell division, just like the canonical version. Another isoform, derived from the PTEN gene, can enter the nucleus, unlike the canonical one that remains in the cytoplasm. These changes in subcellular localization also lead to functional changes," continues Yago Arribas De Sandoval.
Transposable elements and evolution
The researchers then studied the evolutionary pressures exerted on these isoforms in order to understand how the transposable elements influence the diversity of genes during evolution.
In collaboration with Prof. Lluís Quintana-Murci of the Institut Pasteur, they observed that transposable elements are preferentially found in ancient genes, common to all living beings. In addition, they found that the expression of isoforms containing transposable elements increases over time, becoming more conserved and suggesting that they acquire increasing importance for the cell. It is therefore an engine for the diversity of proteins that exist in the cell, allowing it to select the most advantageous isoforms.
"Although transposable elements are already considered as a reservoir of new sequences, this is the first time that such a process has been systematically characterized, combining evolutionary and proteomic studies. Beyond the evolutionary implications, we describe more than a thousand new protein isoforms never recorded, opening new perspectives for the study of cellular functions, in physiological and pathological conditions," concludes Yago Arribas De Sandoval.
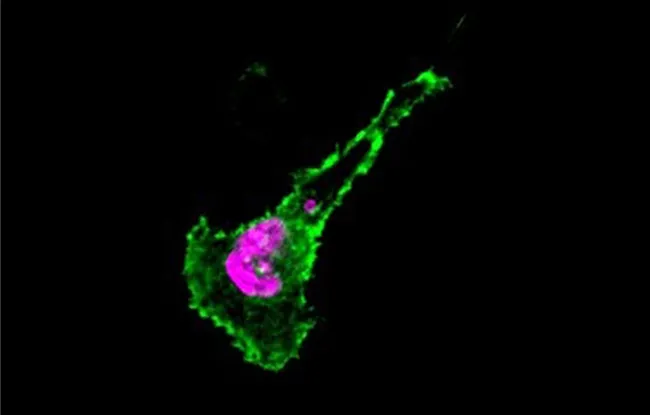
Figure: Confocal microscopy image of a novel isoform of IL15RA (in green), expressed on the cell surface. The cell nucleus is highlighted in pink.
Research News
Discover all our news
Celebration
The Immunity and Cancer research unit (U932) celebrates its twentieth anniversary
12/12/2025
Artificial Intelligence
08/12/2025