Presentation

Adaptive immune responses are initiated by the clonal selection of naive T lymphocytes, which recognize their cognate ligands through antigen-specific T cell receptors. T cell receptors are specific for MHC/peptides complexes. These peptides are generated from internalized antigens that are degraded in endocytic compartments.
The peptides are then loaded on MHC molecules and the complexes are transported to the cell surface. Clonal selection also requires the physical encounter of dendritic cells and T lymphocytes, an event that occurs in the T cell zones of lymph nodes. Naïve T cells then expand and differentiate into effector T lymphocytes that acquire either cytotoxic activity or secrete high amounts of certain cytokines. Some of these T cells become “effectos” (the ones that have direct roles in the immune response, including helper functions for CD4+ T cells, and cytotoxic functions for CD8+ T cells), while other T cells differentiate into resting memory cells. The latter are longed lived and can be reactivated upon re-infection with the same microbe, initiating a more rapide and effective recall response.
Our team is interested in the molecular analysis of immune responses, particularly in the context of cancer. Our main objectives are:
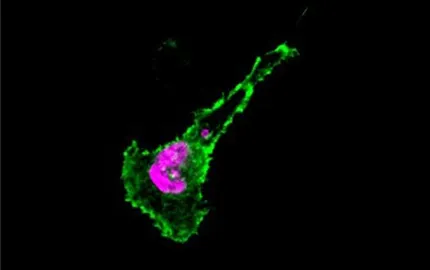
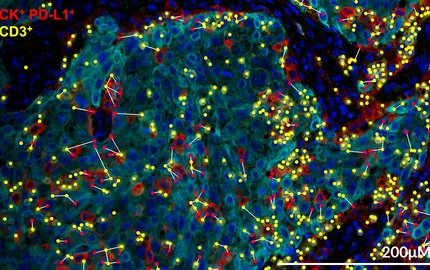
- To understand the molecular basis of antigen presentation in dendritic cells. We analyse the intracellular membrane transport pathways involved and attempt to identify the intracellular compartments where peptide-MHC complexes form. We also investigate the ability and involvement of different mouse and human DC population in immune responses against tumors.
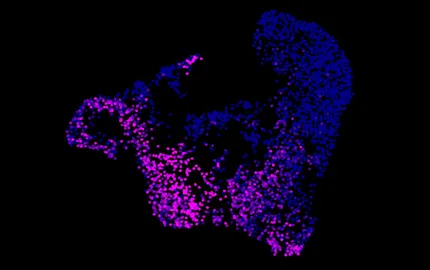
- To analyse the molecular determinants that control gene expression during T cell differentiation and the generation of immunological memory. We attempt to understand how the organisation of chromatin determines the epigenetic control of T cell responses in vitro and in vivo.
Our technological expertise ranges from the most fundamental approaches to study membrane transport in lymphocytes and dendritic cells (subcellular compartmentalization, intravital microscopy, phagosomal functions), the systematic analysis of gene expression and it regulation (RNAseq, Chip Seq, proteomics) and physiological and pathological immune responses (mouse models for cancer immunity, immunomodulation/vaccination, human clinical studies in cancer).