Presentation

My laboratory aims to understand how chromosomal inheritance is achieved with such fidelity in mammalian cells. We study the genetic and epigenetic mechanisms that control the faithful transmission of our genetic material.
Specifically, we are interested in centromeres, key chromosomal loci required for cell division. We aim to identify: i) how centromeres are established, ii) the mechanisms that govern centromere function, iii) how their integrity is maintained during the cell cycle and iv) the role that centromere failure plays in genome instability.
We use molecular and cell biological approaches combined with genetics, physics and biochemistry.
Twitter: @FachinettiLab
Bluesky: dfachinetti.bsky.social
Maintenance of genome integrity and of a correct chromosome karyotype is crucial for human health. Indeed, both an abnormal number of chromosomes – a feature called numerical aneuploidy – and structural chromosome alterations are hallmarks of many human cancers.
Genome instability can arise throughout the whole cell cycle, at crucial moments such as genome replication or cell division. Coordinating faithful chromosome duplication with chromosome segregation is an essential task to maintain a correct karyotype. The Fachinetti laboratory is interested in revealing the processes governing DNA replication and cell division, and how they are coordinated. This is paramount to understand how to prevent the onset of genome instability and consequent pathological states.
Replication stress is a major threat to the integrity of the genome and plays an important causative role in cancer and aging. Replication stress-dependent chromosome breakage is known to target genomic regions that harbor specific features, such late replicating regions, sites of reduced DNA replication initiation, regions that harbor high levels of transcription, or long stretches of repetitive DNA as they are more prone to the formation of DNA secondary structures. Hence, understanding the nature of replication-related lesions and how cells react to replication stress is crucial to elucidate the early events leading to tumorigenesis, as well as for developing novel strategies in anti-cancer therapy.
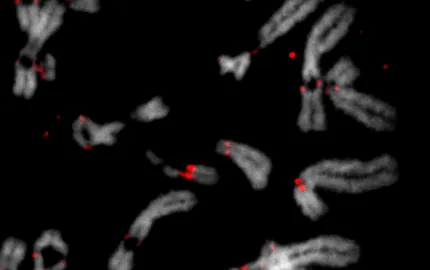
Cell division is an essential stage in the life of all cells. When a cell separates it needs to equally distribute its chromosomes, the molecular basis of genetic heredity, between its two daughter cells (Figure). The centromere plays a key role in this process: it is a small region on each chromosome that serves as an anchor point for the attachment of the cell machinery (the kinetochore) that delivers one copy of each duplicated chromosome to each daughter cell (Mellone and Fachinetti, Cur Biol, 2021). Failure in these processes can lead to chromosome mis-segregation and, consequently, to numerical and structural alterations.
Figure: The centromeres are required for chromosome segregation during both mitosis and meiosis. Immunofluorescence on human cells shows the localization of CENP-C and alpha-tubulin during the different stages of mitosis.
Video: Time lapse video of a human diploid, non-transformed retinal pigment ephitilial-1 (RPE-1) cell during mitosis without (left) or with mis-segregation events (right). DNA is in cyan and alpha-tubulin is in red.
The Fachinetti laboratory aims to identify: i) how centromeres are established, ii) the mechanisms that drive centromere function, iii) how they are replicated and how their integrity is preserved across the cell cycle and iv) the role that centromere failure plays in genome instability. We pay particular attention to the role of centromeric DNA: all native human centromeres are made of repetitive DNA sequences that span several megabases and make up about 3% of the total human genome. We hypothesize that centromeric DNA is a key element of human centromeres to maintain centromere position and assembly, that in turn sustains faithful chromosome segregation (Dumont, M. & Fachinetti, D. PMSB, 2017; Scelfo, A. & Fachinetti, D. Cells, 2019; Gamba, R. & Fachinetti, D. ECR, 2020). On the other hand, centromeric repeats might act as potential fragile regions that, in certain conditions, are more prone to rupture and recombination events (Barra, V. & Fachinetti, D. Nat Com, 2018). Our research program is based on an integrated approach that combines the use of engineered culture models (human cells and mouse embryonic stem cells) to conditionally control the stability of endogenous proteins via an auxin-inducible degron (AID) (Holland, A. J., Fachinetti, D. et al PNAS, 2012; Hoffmann, S. & Fachinetti, D. Meth, 2018) with cell imaging, cytogenetic analysis, DNA sequencing, locus-specific proteomic approaches, genome-wide screening and single molecule technologies on DNA and on chromatin in living cells (Keizer, V et al, BioRxiv, 2021).
We believe our research will provide a transformative view on the molecular mechanisms behind the genesis of genome instability.
References
- Mellone, B* and Fachinetti, D*. (*Co-corresponding authors). Diverse mechanisms of centromere specification. (2021) Curr Biol. Nov 22;31(22):R1491-R1504. https://pubmed.ncbi.nlm.nih.gov/34813757/
- Dumont, M and Fachinetti, D (2017). DNA sequences in centromere formation and function. Book chapter Springer, Centromeres and Kinetochores. Prog Mol Subcell Biol. 56:305-336. https://pubmed.ncbi.nlm.nih.gov/32944612/
- Scelfo, A. and Fachinetti, D. (2019). Keeping the centromere under control: a promising role for DNA methylation. Cells, 16;8(8). https://pubmed.ncbi.nlm.nih.gov/31426433/
- Gamba, R. and Fachinetti, D. From evolution to function: two sides of the same CENP-B coin? (2020) Exp Cell Res. May 15;390(2):111959. https://pubmed.ncbi.nlm.nih.gov/32173469/
- Barra, V. and Fachinetti, D. (2018). The dark side of the centromeres: types, causes and consequences of structural abnormalities implicating centromeric DNA. Nature Communications, 9(1):4340. https://pubmed.ncbi.nlm.nih.gov/30337534/
- Holland, A.J.*, Fachinetti, D.*, Han, J.S., and Cleveland, D.W. (2012). (* Co-first authorship). Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. PNAS, 109, E3350-7. https://pubmed.ncbi.nlm.nih.gov/23150568/
- Hoffmann, S and Fachinetti, D (2018). Real-Time De Novo Deposition of Centromeric Histone-Associated Proteins Using the Auxin-Inducible Degradation System. Book chapter, Methods Mol Biol. 2018;1832:223-241. https://pubmed.ncbi.nlm.nih.gov/30073530/
- Gamba, R., Mazzucco, G., Wilhelm, T., Chardon, F., Velikovsky, L., Picotto, J., Doksani, Y. and Fachinetti, D. A method to enrich and purify centromeric DNA from human cellsbioRxiv 2021.09.24.461328; doi: https://doi.org/10.1101/2021.09.24.461328
- Keizer, V., Grosse-Holz, S., Woringer, M., Zambon, L., Aizel, K., Bongaerts, M., Kolar-Znika, L., Scolari, V., Hoffmann, S., Banigan, E., Mirny, L.A., Dahan, M., Fachinetti, D.* and Coulon, A. * (*Co-corresponding authors). Live-cell micromanipulation of a genomic locus reveals interphase chromatin mechanics. bioRxiv 2021.04.20.439763; doi: https://doi.org/10.1101/2021.04.20.439763
























 MEET THE PI
MEET THE PI