Presentation

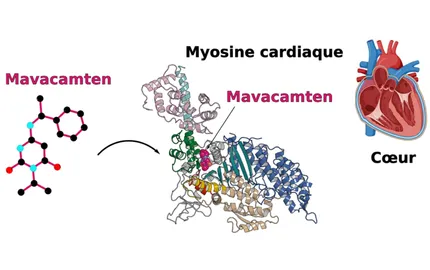
Life is driven by awe-inspiring coordinated movements observed in cells and tissues. In each cell, nm-size molecular motor proteins contribute to these movements as they power numerous mechanical processes with precision and complex orchestration. For the multiple functions that an eukaryotic cell accomplish, motility is essential both at molecular and cellular scales. Our goal is to study the principles underlying how motors convert chemical energy into mechanical movement and how biological motor proteins have evolved to enable distinct cellular function, allowing distinct generation of force precisely activated in space and time. Targeting these nanomotors can be beneficial for human health. Allosteric sites for specific small molecules can act as activators or inhibitors of the force produced by these nanomotors, and we study drugs qualified as breakthrough therapy by the FDA, currently in phase 3 clinical trials against cardiomyophathies. While frequent sites of mutations in these motors can lead to disease phenotypes, high therapeutic potential of allosteric effectors is now established and we aim to extend this knowledge to treat other pathologies, such as cancer and malaria.
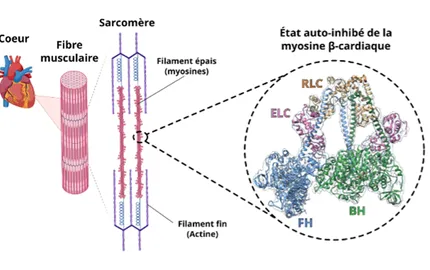
The major focus of the Structural Motility Team is to understand how cytoskeleton nanomotors function by coupling structural biology and functional and cellular assays in order to find insights for their specific cellular roles. Our aim is to understand how interaction of these nanomotors with their actin track can convert chemical energy (supplied by binding and hydrolysis of ATP) into mechanical energy to perform various cell motility processes. Cryo-electron microscopy (Cryo-EM) as well as molecular dynamics contributions have been obtained from our laboratory. This expands the understanding obtained with high-resolution X-ray myosin structures to allow the visualization of the motor in different conformational states. In addition, we are investigating how their activity can be modulated by chemical compounds and future drug candidates. We are also interested in deciphering the exact role they play in cell and how the force generated is controlled.
- Allosteric conformational changes in myosin along the motor cycle and mechanism of force generation.
- Blanc, F., Isabet, T., Benisty, H., Sweeney, H.L., Cecchini, M., Houdusse, A., 2018. An intermediate along the recovery stroke of myosin VI revealed by X-ray crystallography and molecular dynamics. Proc. Natl. Acad. Sci. 115, 6213–6218. https://doi.org/10.1073/pnas.1711512115
- Robert-Paganin, J., Robblee, J.P., Auguin, D., Blake, T.C.A., Bookwalter, C.S., Krementsova, E.B., Moussaoui, D., Previs, M.J., Jousset, G., Baum, J., Trybus, K.M., Houdusse, A., 2019. Plasmodium myosin A drives parasite invasion by an atypical force generating mechanism. Nat. Commun. 10, 3286. https://doi.org/10.1038/s41467-019-11120-0
- Robert-Paganin, J., Pylypenko, O., Kikuti, C., Sweeney, H.L., Houdusse, A., 2020. Force Generation by Myosin Motors: A Structural Perspective. Chem. Rev. 120, 5–35. https://doi.org/10.1021/acs.chemrev.9b00264
- Robert-Paganin, J., Xu, X.-P., Swift, M.F., Auguin, D., Robblee, J.P., Lu, H., Fagnant, P.M., Krementsova, E.B., Trybus, K.M., Houdusse, A., Volkmann, N., Hanein, D., 2021. The actomyosin interface contains an evolutionary conserved core and an ancillary interface involved in specificity. Nat. Commun. 12, 1892. https://doi.org/10.1038/s41467-021-22093-4
- Pospich, S., Sweeney, H.L., Houdusse, A., Raunser, S., 2021. High-resolution structures of the actomyosin-V complex in three nucleotide states provide insights into the force generation mechanism. eLife 10, e73724. https://doi.org/10.7554/eLife.73724
- Grinzato, A., Auguin, D., Kikuti, C., Nandwani, N., Moussaoui, D., Pathak, D., Kandiah, E., Ruppel, K.M., Spudich, J.A., Houdusse, A., Robert-Paganin, J., 2023. Cryo-EM structure of the folded-back state of human β-cardiac myosin. Nat. Commun. 14, 3166. https://doi.org/10.1038/s41467-023-38698-w
How mutations impair myosin function and how small molecules may tune myosin force generation and lead to novel therapies.
Planelles-Herrero, V.J., Hartman, J.J., Robert-Paganin, J., Malik, F.I., Houdusse, A., 2017. Mechanistic and structural basis for activation of cardiac myosin force production by omecamtiv mecarbil. Nat. Commun. 8, 190. https://doi.org/10.1038/s41467-017-00176-5
Robert-Paganin, J., Auguin, D., Houdusse, A., 2018. Hypertrophic cardiomyopathy disease results from disparate impairments of cardiac myosin function and auto-inhibition. Nat. Commun. 9, 4019. https://doi.org/10.1038/s41467-018-06191-4
Moussaoui, D., Robblee, J.P., Robert-Paganin, J., Auguin, D., Fisher, F., Fagnant, P.M., Macfarlane, J.E., Schaletzky, J., Wehri, E., Mueller-Dieckmann, C., Baum, J., Trybus, K.M., Houdusse, A., 2023. Mechanism of small molecule inhibition of Plasmodium falciparum myosin A informs antimalarial drug design. Nat. Commun. 14, 3463. https://doi.org/10.1038/s41467-023-38976-7
Gyimesi, M., Horváth, Á.I., Túrós, D., Suthar, S.K., Pénzes, M., Kurdi, C., Canon, L., Kikuti, C., Ruppel, K.M., Trivedi, D.V., Spudich, J.A., Lőrincz, I., Rauscher, A.Á., Kovács, M., Pál, E., Komoly, S., Houdusse, A., Málnási-Csizmadia, A., 2020. Single Residue Variation in Skeletal Muscle Myosin Enables Direct and Selective Drug Targeting for Spasticity and Muscle Stiffness. Cell 183, 335-346.e13. https://doi.org/10.1016/j.cell.2020.08.050
Hartman, J.J., Hwee, D.T., Robert-Paganin, J., Chuang, C., Chin, E.R., Edell, S., Lee, K.H., Madhvani, R., Paliwal, P., Pernier, J., Sarkar, S.S., Schaletzky, J., Schauer, K., Taheri, K.D., Wang, J., Wehri, E., Wu, Y., Houdusse, A., Morgan, B.P., Malik, F.I., 2024. Aficamten is a small-molecule cardiac myosin inhibitor designed to treat hypertrophic cardiomyopathy. Nat. Cardiovasc. Res. 3, 1003–1016. https://doi.org/10.1038/s44161-024-00505-0
Auguin, D., Robert-Paganin, J., Réty, S., Kikuti, C., David, A., Theumer, G., Schmidt, A.W., Knölker, H.-J., Houdusse, A., 2024. Omecamtiv mecarbil and Mavacamten target the same myosin pocket despite opposite effects in heart contraction. Nat. Commun. 15, 4885. https://doi.org/10.1038/s41467-024-47587-9
Radnai, L., Young, E.J., Kikuti, C., Toth, K., Zhou, M., Hafenbreidel, M., Stremel, R.F., Lin, L., Pasetto, P., Jin, X., Patel, A., Conlon, M., Briggs, S.B., Heidsieck, L., Sweeney, H.L., Sellers, J., Krieger-Burke, T., Martin, W.H., Sisco, J., Young, S., Pearson, P., Rumbaugh, G., Araldi, G.L., Duddy, S.K., Cameron, M.D., Surman, M., Houdusse, A., Griffin, P.R., Kamenecka, T.M., Miller, C.A., 2025. Development of clinically viable non-muscle myosin II small molecule inhibitors. Cell 188, 4604-4621.e15. https://doi.org/10.1016/j.cell.2025.06.006
- Myosin adaptations to specific cell biology roles.
- Ropars, V., Yang, Z., Isabet, T., Blanc, F., Zhou, K., Lin, T., Liu, X., Hissier, P., Samazan, F., Amigues, B., Yang, E.D., Park, H., Pylypenko, O., Cecchini, M., Sindelar, C.V., Sweeney, H.L., Houdusse, A., 2016. The myosin X motor is optimized for movement on actin bundles. Nat. Commun. 7, 12456. https://doi.org/10.1038/ncomms12456
- Pylypenko, O., Welz, T., Tittel, J., Kollmar, M., Chardon, F., Malherbe, G., Weiss, S., Michel, C.I.L., Samol-Wolf, A., Grasskamp, A.T., Hume, A., Goud, B., Baron, B., England, P., Titus, M.A., Schwille, P., Weidemann, T., Houdusse, A., Kerkhoff, E., 2016. Coordinated recruitment of Spir actin nucleators and myosin V motors to Rab11 vesicle membranes. eLife 5, e17523. https://doi.org/10.7554/eLife.17523
- Yu, I.-M., Planelles-Herrero, V.J., Sourigues, Y., Moussaoui, D., Sirkia, H., Kikuti, C., Stroebel, D., Titus, M.A., Houdusse, A., 2017. Myosin 7 and its adaptors link cadherins to actin. Nat. Commun. 8, 15864. https://doi.org/10.1038/ncomms15864
- Arthur, A.L., Songster, L.D., Sirkia, H., Bhattacharya, A., Kikuti, C., Borrega, F.P., Houdusse, A., Titus, M.A., 2019. Optimized filopodia formation requires myosin tail domain cooperation. Proc. Natl. Acad. Sci. 116, 22196–22204. https://doi.org/10.1073/pnas.1901527116
- Houdusse, A., Titus, M.A., 2021. The many roles of myosins in filopodia, microvilli and stereocilia. Curr. Biol. CB 31, R586–R602. https://doi.org/10.1016/j.cub.2021.04.005
- Canon, L., Kikuti, C., Planelles-Herrero, V.J., Lin, T., Mayeux, F., Sirkia, H., Lee, Y.I., Heidsieck, L., Velikovsky, L., David, A., Liu, X., Moussaoui, D., Forest, E., Höök, P., Petersen, K.J., Morgan, T.E., Di Cicco, A., Sirés-Campos, J., Derivery, E., Lévy, D., Delevoye, C., Sweeney, H.L., Houdusse, A., 2023. How myosin VI traps its off-state, is activated and dimerizes. Nat. Commun. 14, 6732. https://doi.org/10.1038/s41467-023-42376-2