- Home >
- Units research >
- Genome Integrity, RNA and Cancer (UMR3348)
Presentation

1. Overview
The research conducted at the « Genome Integrity, RNA and Cancer » Unit explores multiple aspects of genome dynamics that integrate RNA biology in the context of genome maintenance and expression in normal and pathological situations, including cancer.
The Unit has a broad interest in understanding the molecular mechanisms underlying genome integrity and expression, from their basic functioning up to their pathological deregulation in human diseases such as cancer. By bringing together a unique complementarity of scientific and technical expertise, ranging from DNA replication, repair, and recombination, dynamics of gene expression, RNA biology, cancer immunology and cytoskeletal functions, one of our major goal will be to pursue our efforts to foster intra-Unit collaborations to help understanding at unprecedented levels the relationships between DNA metabolism, RNA metabolism and protein regulation by post-translational modifications. Beyond the complementary research expertise of the teams’ Unit, an added value of the groups gathered at the Unit is the use of a wide range of available technologies/approaches (yeast genetics, biochemistry, mammalian cell-based assays, genetically-modifiable mouse models, tumorgrafts of human cells, genomics) and a diversity in biological models from unicellular eukaryotic organisms up to tumor cells and mice models that will also foster intra-Unit collaborations. In line with the scientific objectives detailed below, the Unit name « Genome integrity, RNA and Cancer” has been chosen to express (i) our long-standing interest in the study of the DNA damage response, (ii) our commitment to add the “RNA dimension” in our understanding of genome integrity and, (iii) our willingness to translate our findings to the clinics, particularly in the context of cancer. For all these aspects, both basic and translational, our Unit benefits from the scientific and medical environment of Institut Curie. To strengthen our programs, accelerate scientific discovery and opportunities for clinical translation, we tend to always promote interaction and collaboration with other Units at the Institut Curie, the Curie translational department, the Curie technological platforms (CurieCoreTech) and with several departments at the Curie hospital. Overall, we envisage that our distinctive approach based on the integration of complementary research activities of our teams in the field of genome integrity and expression will lead to paradigm shifts and high profile research output as well as long-lasting benefits for human health.
2. Scientific objectives of the Unit
Living organisms are continuously exposed to DNA damage from endogenous (i.e. DNA replication, oxidative stress) or exogenous (i.e. irradiation, chemo-therapy) origins that impact genome integrity and expression. The DNA Damage Response is therefore critical to orchestrate a network of molecular pathways (DNA repair, gene expression, cell cycle control…) that ultimately impact cell fate decision. The main objective of the teams in our Unit is to understand the basic molecular and cellular mechanisms that underlie genome integrity and expression and how the deregulation of these pathways contributes to cancer development and human diseases. Thanks to the complementary expertise of the teams, we can address those questions at multiple scales with an integrative vision of the cell and the organism, from RNA and chromosome biology up to the cytoskeleton control of cell division.
The Unit’s teams work in a joint effort and coordinated manner on the following three common themes:
- DNA replication stress from novel mechanisms to human disease.
- Mutual interactions between the DNA damage response and RNA metabolism.
- Cytoskeletal functions in chromosome segregation
3. Technological objectives
We intend to implement innovative technologies to simultaneously visualize, at the single cell level, up to 50 protein markers in tissue sections of interest using a novel in situ multiplexed imaging approach termed CODEX (CO-detection by InDEXing). This technology is new in Europe and our Unit is the first one in France to have bought the required equipment and to implement it. In addition, in a combined effort with the Orsay microscopy facility, we intend to implement in situ transcriptomics to enable analysis of cellular transcriptional states with spatial resolution, in tissue samples. This will allow us to interrogate specific pathways in tissue samples at the RNA level.
4. Translational objectives
Besides our basic research activities, several teams investigate translational aspects that can derive in new treatments for cancer (i.e. breast cancer, T-ALL, melanoma). The Unit continuously promotes the interactions with industrial partners and clinicians from the Curie hospital, but also from other hospitals in the Paris area (i.e. Gustave Roussy).
5. Teaching
The Unit has contributed to teaching and training at multiple levels. All group leaders and senior researchers are involved in hosting students, organizing and/or teaching Masters Courses. Together with the two Curie international courses (Curie training) organized by other Curie Units on “Non-coding genome” and “Epigenetics”, the two Curie international courses organized by our Unit on “Genome instability and human diseases” and “Post-transcriptional gene expression” form an interesting series of courses on the general topic of “Genome Dynamics”..
6. Summary of the recent work of our teams
Team “Controlling microtubule dynamics and function with the tubulin code” (Carsten JANKE)
- Purifying tubulin with controlled posttranslational modifications (Nat Protoc 2019) allowed us to pin down how tubulin polyglutamylation controls microtubule interactions with other proteins (EMBO J 2023). A new lysate-based approach (Nat Cell Biol 2022) now opens the possibility to test a large number of microtubule interacting proteins.
- Deregulation of polyglutamylation causes neurodegeneration and reduced cargo transport in neurons (EMBO J 2018a, 2018b, J Cell Sci 2020). The control of axonal transport as well as neurodegeneration can be attributed to one specific glutamylating enzyme, TTLL1 (EMBO J 2021).
- Tubulin glycylation controls the beat of sperm flagella, and thereby their swimming pattern. Lack of glycylation leads to male subfertility (Science 2021), photoreceptor degeneration (J Cell Sci 2017) and defects of primary cilia (J Cell Biol, 2017) that can promote tumorigenesis (EMBO J, 2014).
Team “DNA recombination during replication for genome stability” (Sarah LAMBERT)
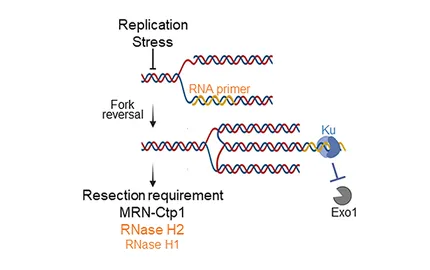
- The maintenance of replication fork integrity and replication competence require a well-orchestrated degradation of nascent strands (Nat Com., 2017, Life Sci Alliance 2019) and a routing towards Nuclear Pores Complexes (Nat Com., 2020) to avoid pathological DNA structures in mitosis (Mol. Cell, 2017).
- The protection of replication forks requires the end joining factor KU and Okazaki fragments that generates RNA:DNA hybrids for KU binding (Mol. Cell 2023).
- Histone deposition at dysfunctionnal fork fine-tunes replication restart and impacts genome stability at replication stress site (PLoS Biology, 2014; PLoS Genetics 2019).
Team “Chromatin and RNA splicing ” (Reini LUCO)
- Cancer cells take advantage of specific changes in RNA splicing during a cell reprogramming called epithelial-to-mesenchymal transition (EMT) to gain new phenotypic traits essential for cell migration, invasion and resistance to treatment (BMC Biol, 2021).
- These splicing changes are orchestrated by dynamic changes in histone marks that coordinate functionally related splicing events (Nature Str. Mol. Biol, 2015; Nat Comm, 2021)
- Highly localized changes in these splicing-associated histone marks using CRISPR/dCas9 epigenomic editing are sufficient to induce a splicing change that will favor the EMT phenotype, supporting its use in therapy (Cell Rep, 2022).
Team “RNA, tumor microenvironment and metastasis” (Albertas NAVICKAS)
- RNA-binding proteins control mRNA regulons implicated in cancer metastasis: HNRNPC links alternative polyadenylation to translational control (Navickas, Asgharian et al., 2023, Nat Cell Bio), SNRPA1 and TARBP2 recognise RNA structural elements and impact target mRNA splicing (Fish, Khoroshkin, Navickas et al., 2021, Science; Fish, Navickas et al., 2019, Mol Cell), RBMS1 controls mRNA stability (Yu, Navickas, Asgharian et al., 2020, Cancer Discov).
- human stem cell-derived lung organoids model the infection by SARS-CoV-2 and reveal the role of androgen signalling in the expression of SARS-CoV-2 receptors (Samuel, et al., 2020, Cell Stem Cell).
Team “RNA biology, signaling and cancer” (Stéphan VAGNER)
- DNA-damage response RNA-binding proteins. (DD-RBPs) control multiple aspects of the DNA damage response, from (post-)transcriptional gene expression to DNA repair (TIBS 2014, J. Mol. Biol., 2017a, 2017b, NAR 2020, TIBS 2021, EMBO 2023).
- Targeting the eIF4F translation initiation complex relieves the resistance to anti-cancer therapies (Nature, 2014; Cancer Res., 2016, 2017, Nature Med., 2018, Nat Com., 2019, Cell 2020; Nat. Rev. Cancer, 2021).
- Identification of molecular pathways critical to Acute Lymphoblastic Leukemia (ALL) progression (Cancer Cell 2015; Leukemia 2016; PLoSOne 2021; Sci Transla Med 2021; Cancer Discov 2021) and demonstration that targeting the T cell receptor (TCR) complex with clinically-relevant anti-CD3 mAbs is anti-leukemic and highly cooperative with chemotherapy in T-ALL xenografts (Cancer Discov 2016; Blood 2020).