Presentation

DNA replication must adapt to changes in chromatin organization associated with cell differentiation and development, whose deregulation can challenge genome stability and leads to mutations, cancer and many other genetic diseases. However, despite intensive studies, the mechanisms that coordinate where and when replication initiates in the human genome remain poorly known. Our team focuses on using cutting-edge high-throughput genomic approaches and genome-wide data analyses to study the spatio-temporal replication program of the human genome and its impact on genome stability, in particular to address the following questions:
- What determines the replication program, i.e. the position, the time of firing and the efficiency of replication origins in the human genome?
- How this program is regulated and associated with gene transcription and chromatin organization?
- How deregulation of these programs challenges genome stability and leads to human diseases?
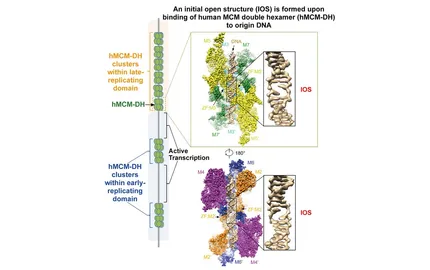
In collaboration with experimental biologists, we have developed a method (Repli-Seq) and generated one of the first high-resolution replication timing profiles of the human genome (Fig. 1). Studies of these profiles from different human cell types have allowed us to reveal that the genome is organized in megabase replication domains associated with higher order chromatin structural units. By evolutionary analyses, we have also established that replication is a major process driving genome mutational landscape in normal and cancer cells. We are now applying Repli-Seq technique to analyze the replication dynamics from cells upon replication stress to study how deregulation of the replication program challenges genome stability, in particular, common fragile site activity (Fig. 2).
More recently, we developed a new tool using machine learning algorithms. MnM automates the analysis of this temporal parameter of DNA replication. Based on data generated from over 119,000 human cells, the authors show that heterogeneous subpopulations of cells can be distinguished, paving the way for a better understanding of the mechanisms involved in the development of physiological and pathological tissues.

Figure 3: Copy number variations (CNVs) of single cells are used by a deep learning model to identify the replication state of cells. Cells that are not in the replication phase are then clustered using UMAP and DBSCAN algorithms to discover the underlying subpopulations. Finally, cells in the replication phase are associated with these subpopulations, allowing the reconstruction of three distinct subpopulations based on replication time, as illustrated by the CNV profiles. This approach highlights genomic heterogeneity in tumor tissues, highlighting somatic copy number alterations and the ubiquitous aneuploidy process during tumorigenesis.
Moreover, we have developed new genome-wide approaches to study replication program at single molecule/cell resolution (Fig. 3), in order to further study the intrinsic (between alleles) and extrinsic (cell to cell) variation in replication and to further investigate the relation between cell-to-cell heterogeneity of replication and the cell-to-cell heterogeneity in gene transcription and chromatin organization.
Our study on DNA replication and genome instability will provide the important bases for further understanding its role during development and aging, and how its deregulation contributes to tumorigenesis and to human diseases.
GitHub page of the team: https://github.com/CL-CHEN-Lab.